CGC 2020 Webinars
Welcome to the webinar series sponsored by the Cancer Genomics Consortium and the University of Wisconsin Collaborative Genomics Conference.
These lecture series focus on topics of high interest for CGC members and attendees of the UW Collaborative Genomic Conference, including implementation of new technologies in clinical genetic/genomic testing, standards and resources for interpretation of sequence and copy number variants, germline predisposition to cancer and novel approaches for detection of structural variants in constitutional and cancer samples.
Circulating tumor DNA testing in oncology: established and emerging approaches, clinical utility and applications
Trevor Pugh, PhD, FACMG
Princess Margaret Cancer Centre, Ontario Institute for Cancer Research, University of Toronto
Clinical use of circulating tumor DNA (ctDNA) testing is rapidly increasing, but this type of analysis remains technically challenging, and its clinical utility and optimal utilization in clinical management of cancer patients are still being investigated. In this webinar, Dr. Pugh will review technical aspects of ctDNA testing (including specimen types and collection, transport, DNA isolation, assay design and performance characteristics); different types of biomarkers that are being monitored by ctDNA approaches (sequence variants, copy number changes, methylation profiles and ctDNA fragmentation patterns); evidence of clinical validity and clinical utility; and applications of ctDNA testing in genomic characterization of tumors at diagnosis, in non-invasive monitoring of advanced cancers, in detection of residual disease and relapse, and even in screening for cancer in asymptomatic individuals.
Resources for Interpreting Cancer Genomes: From Knowledgebases to Gene Lists
Beth Pitel, M.S., CG (ASCP)
Mayo Clinic
Interpreting cancer genomes is a challenging task requiring the use of references from the literature and other helpful resources and tools. In this webinar we will learn about several available databases and knowledge bases that can aid in cancer variant interpretation. Beth Pitel will illustrate how these resources helped the Genomics of Oncology Annotation Team (GOAT) at Mayo Clinic create disease-specific gene lists used as resources for the interpretation of chromosomal microarrays. Additionally, the CGC and the Mayo GOAT are collaborating in an effort to fine-tune a process for curating disease-specific gene lists.
Copy Number Abnormality Detection from Whole Genome Sequencing Data
Alka Chaubey, PhD, FACMG
Head of Cytogenomics, Perkin Elmer Genomics
Dr. Alka Chaubey will discuss the utilization of low pass genome sequencing as a new diagnostic approach for CNV detection across the entire genome. Dr. Chaubey will provide evidence from internal validation studies for CNGnome® test, which by utilizing 5x genome sequencing delivers an increased level of CNV detection when compared to traditional methods. Multiple case examples will be discussed in the webinar to demonstrate the clinical capabilities of CNGnome®. Several examples of whole genome sequencing (>30X depth) with pathogenic sequencing and copy number variants will also be discussed.
Interpretation of Copy Number Abnormalities (CNAs) and CN-LOH in Cancer using the ACMG / CGC Standards: Working through Complex Cases
This webinar provides practical guidance how to streamline interpretation of CNAs and CN-LOH in neoplasia using the current ACMG/CGC standards. Easy steps to generate clear, accurate and well organized reports will be illustrated. Using case examples, the speakers will explain challenging concepts including interpretation of genomic complexity, adjustment of ploidy, estimation of the percentage of involved cells and reporting of abnormalities suspected to be germline.

Fady M. Mikhail, MD, PhD, FACMG
Co-Director, Cytogenetics Laboratory
Professor, Department of Genetics
University of Alabama at Birmingham

Gordana Raca, MD, PhD, FACMG
Director of Clinical Cytogenomics, Center for Personalized Medicine,
Department of Pathology
Children's Hospital Los Angeles
Associate Professor, Clinical Pathology, Keck School of Medicine of USC

Vanessa Horner, PhD, FACMG
Director, UW Cytogenetics and Molecular Genetics Services WSLH
Assistant Professor, Department of Pathology and Laboratory Medicine
University of Wisconsin-Madison

Optical Mapping and its Role as a Cytogenomics Tool in Cancer
Dr. Brynn Levy, MSc (Med), PhD, FACMG
Columbia University Medical Center,
New York Presbyterian Hospital
Dr. Levy will describe the principles of optical mapping, illustrate the capacity of this technology to detect balanced structural rearrangements in different sample types, and compare its performance with classic cytogenetic approaches including karyotype analysis and fluorescence in situ hybridization testing.

Application of Optical Mapping for Comprehensive Assessment of Structural Rearrangements in Hematological Malignancies
Dr. Rashmi Kanagal-Shamanna, MD
The University of Texas MD Anderson Cancer Center
Dr. Kanagal-Shamanna will discuss the application of optical mapping in comprehensive genomic characterization of hematological malignancies and highlight the clinical utility of this technique in management of including myelodysplastic syndromes.

Accreditation Statement
In support of improving patient care, the University of Wisconsin Madison ICEP is jointly accredited by the Accreditation Council for Continuing Medical Education (ACCME), the Accreditation Council for Pharmacy Education (ACPE), and the American Nurses Credentialing Center (ANCC) to provide continuing education for the healthcare team.
Credit Designation Statements
The University of Wisconsin Madison ICEP designates this live activity for a maximum of 1.0 AMA PRA Category 1 Credits. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
The University of Wisconsin Madison, as a member of the University Continuing Education Association (UCEA), authorizes this program for .01 continuing education units (CEUs) or 1 hour.
Intended Audience
PhD, MD, RN, Genetic Counselors, and healthcare professionals who generate or use genomic/genetic data in their practice
Global Objectives
As a result of this educational regularly scheduled series, learners will be able to:
1. Recognize both common and rare genetic abnormalities associated with developmental delay, infertility, prenatal ultrasound findings, and oncology specimens.
2. Demonstrate an improved ability to effectively communicate genetic information across healthcare team members from different professions and disciplines.
3. Examine performance and utility of novel testing approaches and standardized classification recommendations.
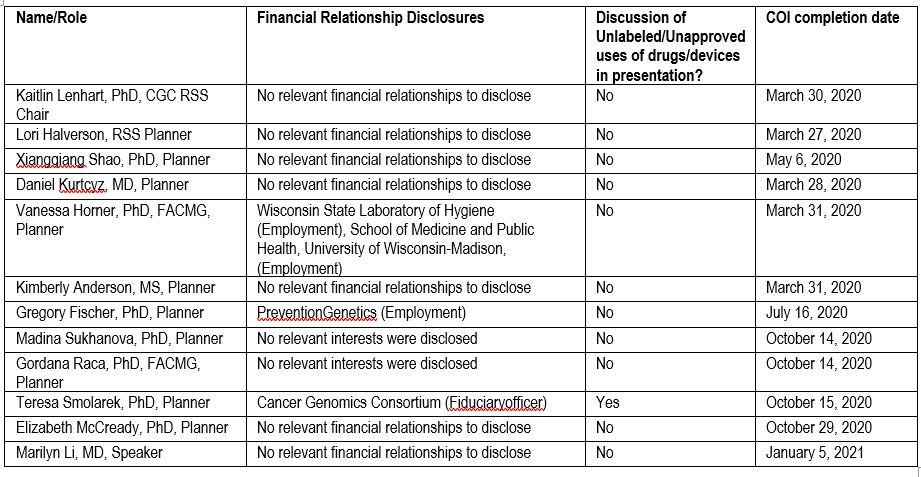
Policy on Disclosure:
It is the policy of the University of Wisconsin-Madison ICEP that the faculty, authors, planners, and other persons who may influence content of this CE activity disclose all relevant financial relationships with commercial interests* in order to allow CE staff to identify and resolve any potential conflicts of interest. Faculty must also disclose any planned discussions of unlabeled/unapproved uses of drugs or devices during their presentation(s). For this educational activity, all conflicts of interest have been resolved and detailed disclosures are listed below.
* The University of Wisconsin-Madison ICEP defines a commercial interest as any entity producing, marketing, re-selling, or distributing health care goods or services consumed by, or used on, patients The University of Wisconsin-Madison ICEP does not consider providers of clinical service directly to patients to be commercial interests.